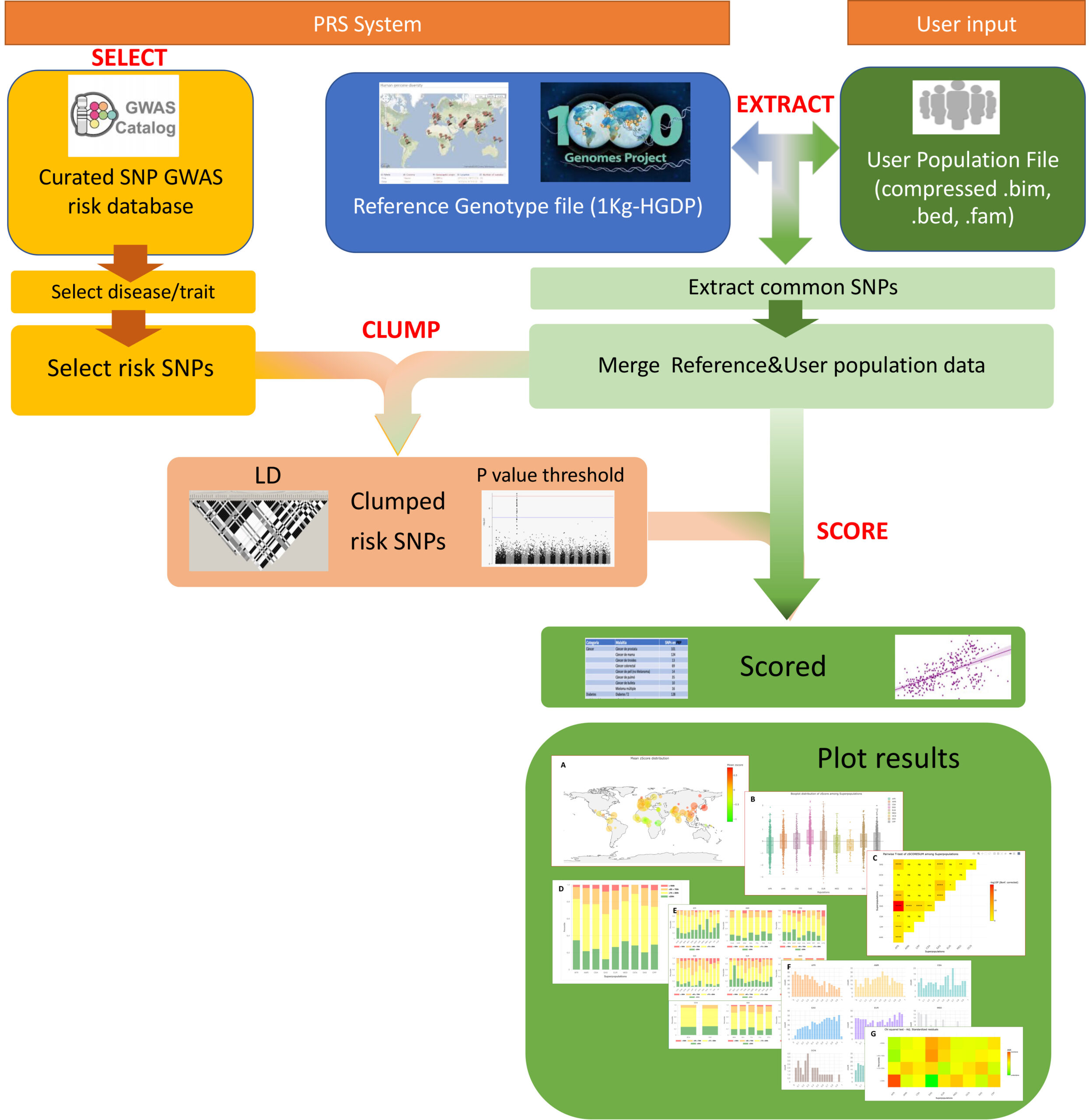
Polygenic risk score comparator (PRScomp)
target population vs. worldwide populations
PRScomp allow users to evaluate polygenic risk score on a own tested population and compare it with worldwide populations from 1000 Genome Project (1KG3) and Human Genome Diversity Project (HGDP)
# To test PRScomp functionality
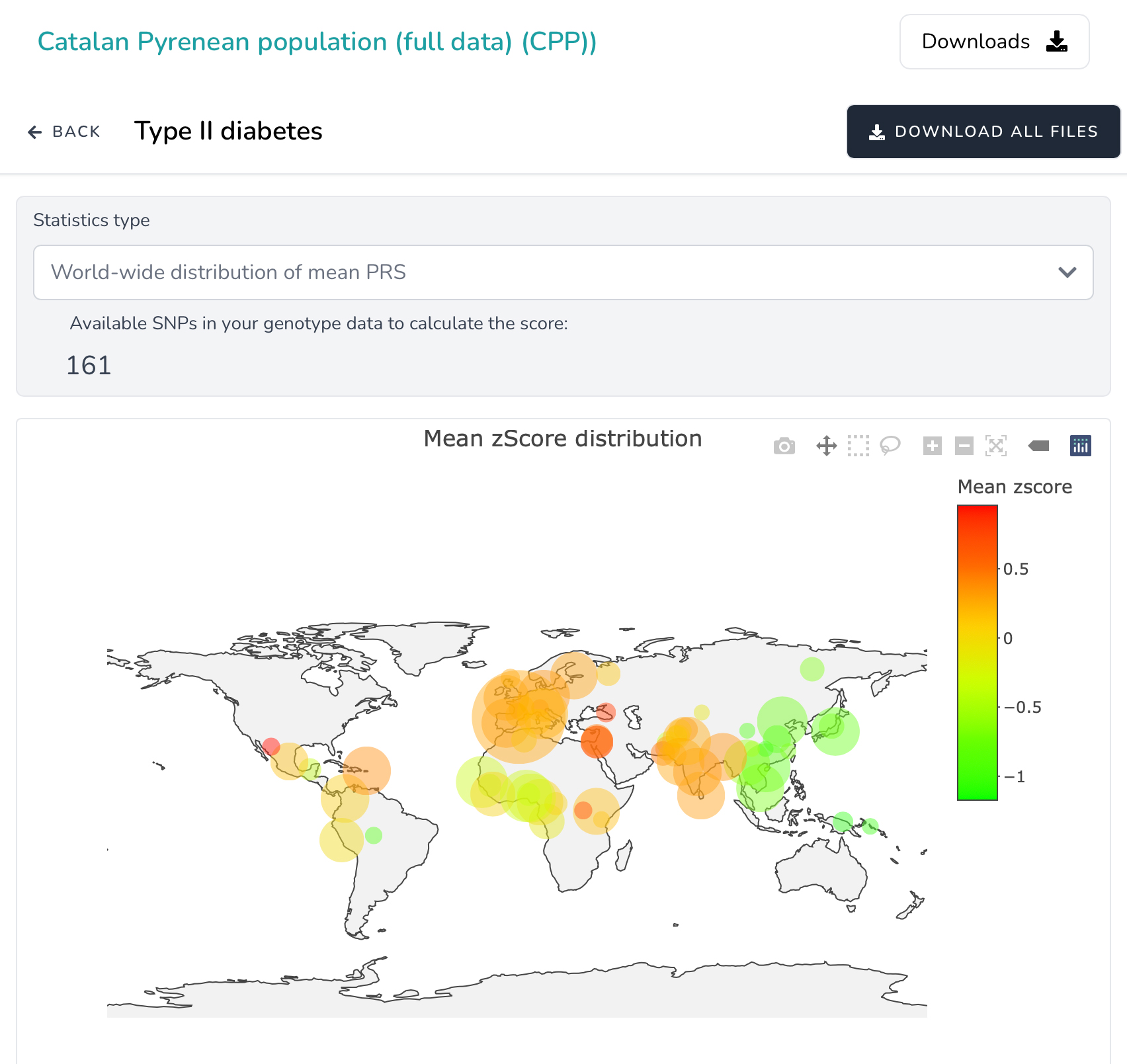
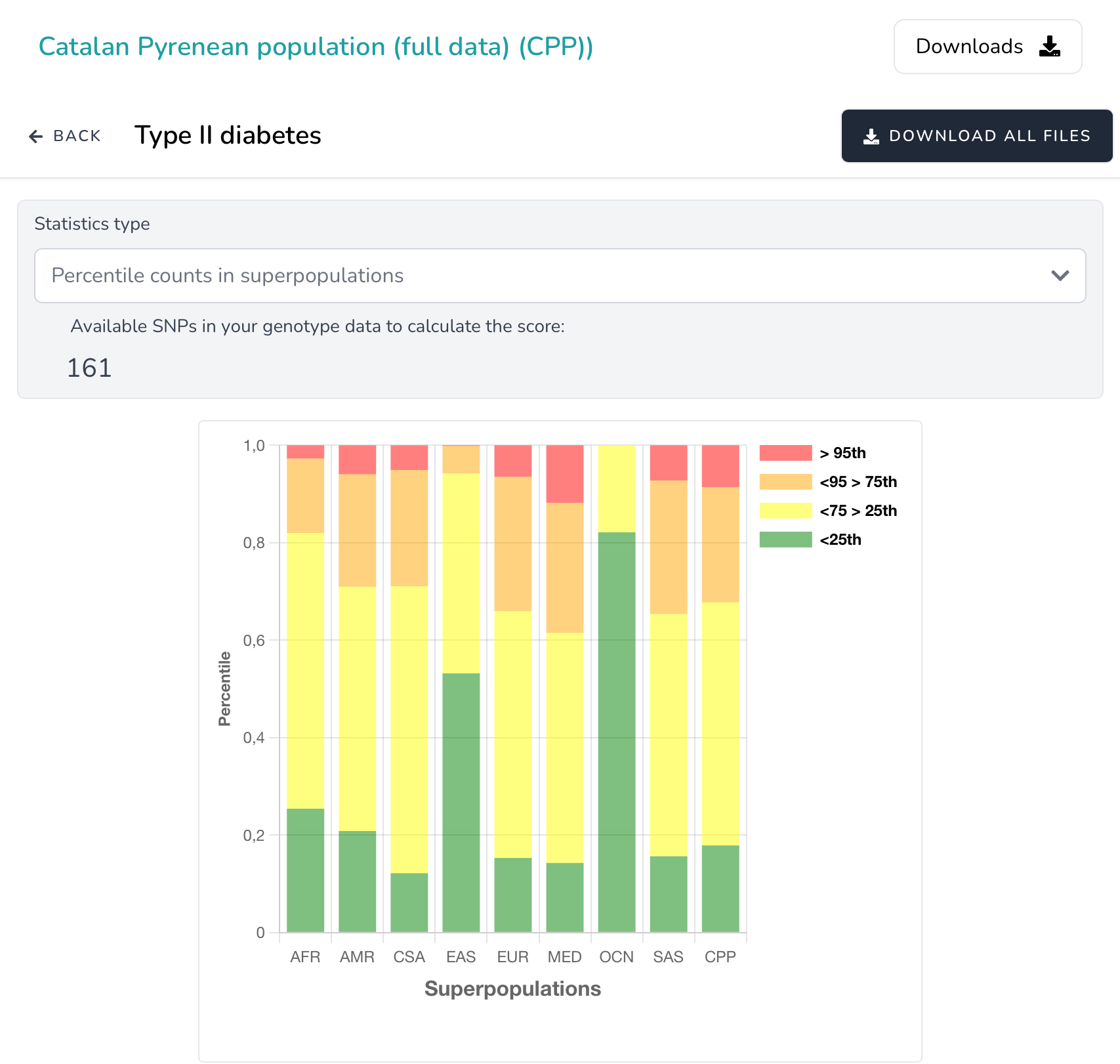
You can test PRScomp functionality on the Catalan Pyrenean population sample of 397 unrelated individuals. First, select a disease/trait of interest (Select disease/trait option). Second, chose the most appropriate studies and third, view polygenic risk score distribution of the Catalan Pyrenean population compared with worldwide populations.
# To analyze yout own genotype data
In order to analyze your own genotype data, you must be registered to the platform. After registering you can access to additional PRScomp options that allow you to evaluate and visualize the distribution of polygenic risk score of your population for several disease/traits. A sample genotype file can be downloaded from https://figshare.com/articles/dataset/example_geno_zip/20051999
# PRScomp pipeline
Compressed binary plink files (.bed/.bim/.fam) with genotype data of target population is uploaded to the platform by the user. The following steps are performed:
- Compressed file is processed to extract matching disease associated SNPs to Reference genotype dataset.
-
Genotype data of disease associated SNPs of tested population is merged with Reference genotype dataset to obtain a merged file that includes only common disease associated SNPs of both datasets. Merged file is also splitted by gender to obtain a merged-male and merged-female files.
The user can select a desired disease/trait from disease/trait database, to be tested on their own population and be compared to the worldwide 1KG3-HGDP populations. A filtering step including population and target gender, should be considered. A list of studies belonging to the selected disease/trait is presented from which the user can select the most appropriate. Markers (SNPs) from selected studies are extracted and processed as follows:
- Selected markers are clumped using the merged file from worldwide population and the tested population. A curated set of risk markers is obtained and used to calculate summatory polygenic risk score (PRS) by PLINK. Values obtained are normalized along all populations by z-score.
- Distribution of z-scored PRS of target and 1KG3-HGDP superpopulations (African, American, East-Asia, Central-Asia, South-Asia, European, Oceanian and Middle-East) are plotted by boxplot. Mean z-scored PRS values of target and worldwide populations are plotted by bobble plot on the worldwide map. Finally, a barplot of percentile distribution of z-scored PRS among target population and 1KG3-HGDP superpopulations is presented.
- Differences on the z-scored PRS distribution among target population and 1KG3-HGDP superpopulations is tested by pairwise t-test, as well as the differences among populations for percentile counts is evaluated by the adjusted standardized residuals.
The user can download graphical representation of results as well as the merged file and raw PRS results in plain text format.
PRScomp uses PLINK binary file as input file. This format file has tree components, the .bed file is a binary biallelic genotype table that include genotype data of subjects, the .bim file include variant information and the .fam file include sample information. Marker coordinates on .bim file must be done in human genome assembly GRCh38 (hg38). Prior to the upload on PRScomp, these three files must be compressed in a single «zip» file.
# PRScomp Database
A disease/trait database is constructed based on GWAS Catalog summary statistics data of genetic risk score of disease and traits. GWAS Catalog SNP records are grouped into a set of query entries by using GWAS Catalog summary statistics fields: “DISEASE/TRAIT”, “MAPPED_TRAIT”, “STUDY ACCESSION”, ancestry population and the unit measure of the SNP effect, extracted from “95% CI (TEXT)” field. Each query entry is assigned to one of these main categories: Cancer/Neoplasm, Disorder/Diseases, Processes/measurements and Trait/conditions. Each category is subdivided into additional subcategories to refine the phenotype under study if necessary. Each query entry includes information on the number of individuals of the study and broad ancestral category of samples. We also provide links to disease/trait, study accession, PubMed ID, Experimental Factor Ontology trait and a link to GWASROCs-Database for those PRScomp entries that are common. As well as, for each associated SNP; SNP rsid, risk allele, risk allele effect (ln(OR), beta) and P-value of association is also included.
SNP were included in the database after performing a manually curated QC. Raw data from GWAS Catalog summary statistics was filtered according to the following criteria:
- Only biallelic single nucleotide variants were considered
- Only SNPs containing information on effect size (OR or BETA) were included
- Only SNPS genotyped at 1KG3 and HGDP (see Reference genotyped data set)
- Ambiguous SNPs (G to C; A to T) were excluded
- SNP with conditional effect were excluded
Worldwide populations analyzed in PRScomp. ACB: African Caribbeans in Barbados; ADY: Adygei; ASW: Americans of African Ancestry in SW USA; BAL: Balochi; BEB: Bengali; BED: Bedouin; BGT: Bergamo Italy; BNK: Bantu Kenya; BOU: Bougainville; BPY: Biaka; BRA: Brahui; BSA: Bantu South Africa; BUR: Burusho; CAM: Cambodian; CDX: Chinese Dai; CEU: European NW (Utah Residents, CEPH) (*); CHB: Han Chinese in Beijing; CHS: Southern Han Chinese; CLM: Colombian Medellin; DAI: Dai; DAU: Daur; DRU: Druze; ESN: Esan Nigeria; FIN: Finnish; FRB: Basque; FRE: French; GBR: British; GIH: Gujarati Indian (from Houston Texas) (*); GWD: Gambian; HAN: Han; HAZ: Hazara; HEZ: Hezhen; IBS: Iberian Spain; ITU: Indian Telugu (from the UK)(*); JAP: Japanese HGDP; JPT: Japanese Tokyo; KAL: Kalash; KAR: Karitiana; KHV: Kinh Vietnam; LAH: Lahu; LWK: Luhya Kenya; MAK: Makrani; MAN: Mandenka; MAY: Maya; MBU: Mbuti Pygmy; MIA: Miao; MON: Mongolian; MOZ: Mozabite; MSL: Mende Sierra Leone; MXL: Mexican; NAX: Naxi; ORC: Orcadian; ORO: Oroqen; PAL: Palestinian; PAP: Papuan; PAT: Pathan; PEL: Peruvians Lima; PIM: Pima; PJL: Punjabi Pakistan; PUR: Puerto Ricans; RUS: Russian; SAN: San; SAR: Sardinian; SHE: She; SIN: Sindhi; STU: Tamil Sri Lankan (from the UK) (*); SUR: Surui; TSI: Tuscan; TU: Tu; TUJ: Tujia; UYG: Uygur; XIB: Xibo; YAK: Yakut; YII: Yi; YRI: Yoruba Ibadan; YUR: Yoruba HGDP. Adapted from Auton et al. 2015 (1KG3) and Bergström et al. 2020 (HGDP). (*) Coordinates based on ancestry location.
Selected markers are clumped using the merged genotype file of worldwide and test populations using PLINK to remove SNPs that are highly correlated but preferentially retaining the SNPs most associated with the phenotype under study. Clumping settings were: P-value threshold for a SNP to be included as an index SNP is set to 1, to ensure that all SNPs are considered (–clump-p1 1), SNPs with r2 higher than 0.1 with the index SNPs will be removed (–clump-r2 0.1), a window of 250 kb of the index SNP is considered (–clump-kb 250) and for each clumped set, the SNP with lowest P value is included (–clump-field P).
The default formula for PRS calculation follows PLINK (1) scoring routine as:
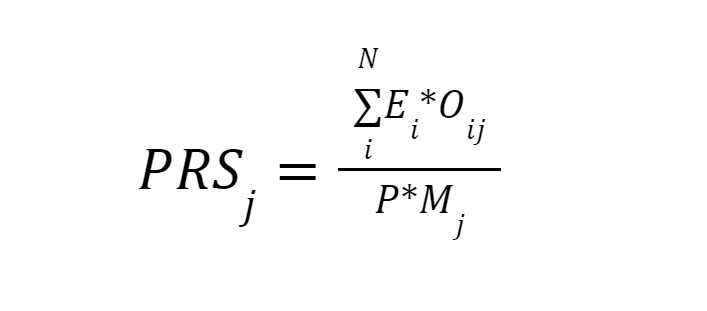
- the effect size of SNP i is Ei;
- the number of effect alleles observed in sample j is Oij;
- the ploidy of the sample is P (2 for autosomes);
- the total number of SNPs included in the PRS is N;
- the number of non-missing SNPs observed in sample j is Mj.
1 PLINK SNP scoring routine at https://zzz.bwh.harvard.edu/plink/profile.shtml
# Imputation of target population
# About us
PRScomp has been developed by the Group of Genetics and Complex Diseases with the support of the Diputació de Lleida and the GenPIR Working Team.
To contact PRScomp Team, with queries or comments please email:
Dr. Joan Fibla (joan.fibla@udl.cat)
Dr. Marina Laplana (marina.laplana@udl.cat)
# If you use PRScomp please cite:
- Laplana, M., Lopez-Ortega, R., and Fibla, J. (2024). Polygenic risk score comparator (PRScomp): Test population vs. worldwide populations. Int. J. Méd. Inform. 183, 105333. doi: 10.1016/j.ijmedinf.2023.105333
Supported by: